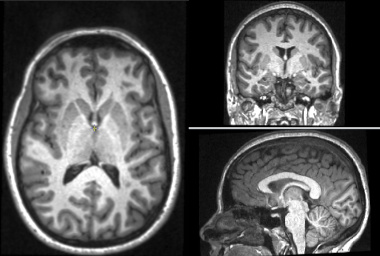
The Brain Atrophy tool can be used to measure brain tissues volumes, or the changes in volume that occur over time. It can separately measure the volumes of grey and white matter, and of the whole brain paranchyma. The tool works with T1-weighted images collected at one or more time-points. In addition, if FLAIR images are available, GM and WM volumes can be assessed in the presence of hyperintense lesional tissue and the lesion volumes computed. Even if hyperintense lesions are not present, use of a FLAIR image may improve tissue classification, although we have not shown this to be the case.
The program uses a template image which is segmented probabilistically into grey-matter, white-matter and CSF. This template is registered to the input T1-weighted images and used as the prior probabilities in an Expectation-Maximization (E-M) segmentation algorithm. If lesions are to be segmented, the segmentation happens after the first E-M classification. A further E-M classification is then performed after excluding the lesions from the GM, WM and CSF tissue classes.
The input images must:

The Brain Atrophy tool is the most computationally demanding of Jim's suite of sub-programs. It requires a lot of both processing power and computer memory. We recommend a system with at least 16 GB of memory, and a modern multi-core processor such as an Intel Core i7 or AMD Ryzen 7 or 9 Even then, you can expect processing times of up to 1 hour per subject if you have multiple time-points to analyse with white-matter lesions.
Start the Brain Atrophy tool from the Toolkits menu:

This will bring up:
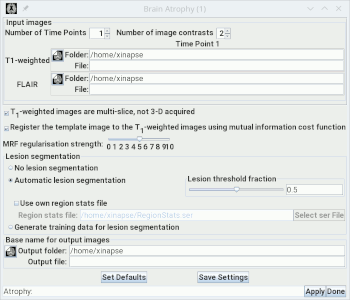
You set the number of time points to be analysed using the
Set the number of image contrasts using the
This will alter the size of the
You can now go on to setup the other
options for analysis.
 spinner. For cross-sectional
analysis, set this to 1. For longitudinal analysis, set this to 2 or more.
spinner. For cross-sectional
analysis, set this to 1. For longitudinal analysis, set this to 2 or more.
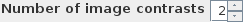 spinner. If you have
just T1-weighted images, set the number of contrasts to 1; if you also want
to use FLAIR images, set the number of contrasts to 2.
spinner. If you have
just T1-weighted images, set the number of contrasts to 1; if you also want
to use FLAIR images, set the number of contrasts to 2.
N Time-Points × M Contrasts matrix of image
selection panels below. For each image selection panel in the matrix, you must select the
appropriate input image (T1-weighted image in the first row, and [optionally]
FLAIR in the second row) corresponding to the time point column.