Brain Atrophy - Cross-Sectional
The picture below shows the setup for assessing brain atrophy at a single time-point
(cross-sectionally) using just a T1-weighted image.
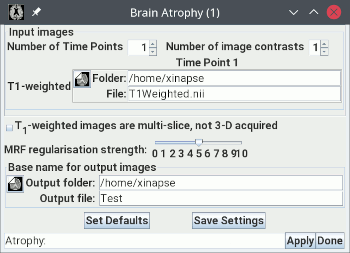
The input image should, ideally, have been acquired using a 3-D (not multi-slice) pulse sequence.
Follow these steps for cross-sectional atrophy assessment:
- Load the T1-weighted image in the input image selection panel. In the
example above, this is called "
T1Weighted.nii".
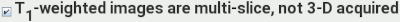 .
If the T1-weighted image was acquired multi-slice (with slice
selection), rather than 3-D (with the slice dimension being
phase-encoded), then select this option.
.
If the T1-weighted image was acquired multi-slice (with slice
selection), rather than 3-D (with the slice dimension being
phase-encoded), then select this option.
- Leave the
 slider at its
default value of 5. This should not need to be changed.
slider at its
default value of 5. This should not need to be changed.
- Set the base name and folder for the output images.
- Optional. If you want to use a FLAIR image as well, increase the number of contrasts
to 2, and select the FLAIR image.
Now press the  button to start the
analysis, which will take some time. When the analysis is complete, you will
see a pop-up message showing the results:
button to start the
analysis, which will take some time. When the analysis is complete, you will
see a pop-up message showing the results:
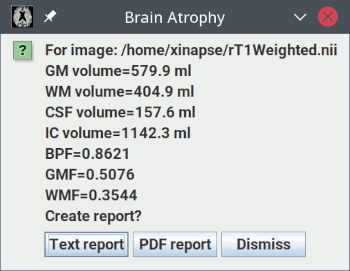
These results show:
- The grey-matter (GM) volume.
- The white-matter (WM) volume.
- The cerebro-spinal fluid (CSF) volume.
- The intra-cranial volume (ICV), which is the sum of the volumes of the 3 compartments
above.
- The brain parenchymal fraction (BPF), which is: (GM Volume + WM Volume) / ICV.
- The grey-matter fraction (GMF), which is: GM Volume / ICV.
- The white-matter fraction (WMF), which is: WM Volume / ICV.
To make a permanent record of these results, you can:
- Write to a text file report. A
File chooser will appear, for you
to choose a log file name. The default file extension for log
files is ".log". If the chosen file already exists, an
entry will be appended to the log file.
- Write to a
PDF file report. A File chooser will appear, for you
to choose a PDF file name. If the chosen file already exists, an
entry will be appended to the PDF file. A PDF report will also include an illustration the
GM/WM/CSF segmentation.
For an input image called "T1Weighted.nii"
and a base name of "Test", these output images will be created in the selected folder:
TestGMPrior.nii - the grey-matter prior probability image, registered to
the T1-weighted image.TestWMPrior.nii - the white-matter prior probability image, registered to
the T1-weighted image.TestCSFPrior.nii - the CSF prior probability image, registered to
the T1-weighted image.TestLVPrior.nii - the lateral ventricles prior probability image, registered to
the T1-weighted image.TestPosition.nii - an image of the pixel positions in
template image space.
rT1Weighted_pGM.nii - the grey-matter posterior probability image.rT1Weighted_pWM.nii - the white-matter posterior probability image.rT1Weighted_pCSF.nii - the CSF posterior probability image.rT1Weighted_pOTHER.nii - the other (non-tissue) class posterior probability
image.rT1Weighted_Classes.nii - a colour image showing the final segmented tissue
classes.
rT1Weighted.nii - a cropped version of the T1-weighted
input image and bcrT1Weighted.nii, a bias-corrected version of this.
The procedure for cross-sectional atrophy assessment is summarised in the flow-chart below.
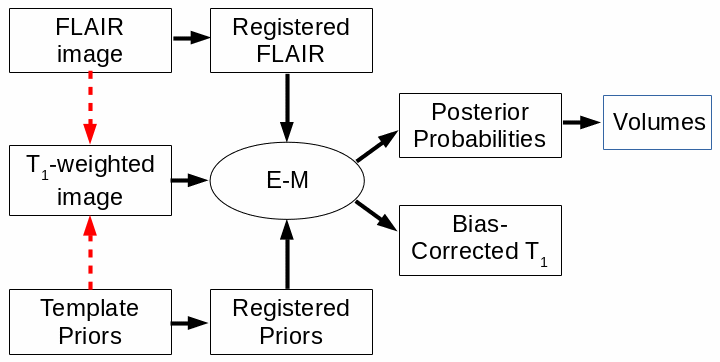
The red arrows indicate a registration step, with the arrow pointing towards the registration
target. The use of the FLAIR image is optional. E-M: expectation-maximization classification
algorithm.


 .
If the T1-weighted image was acquired multi-slice (with slice
selection), rather than 3-D (with the slice dimension being
phase-encoded), then select this option.
.
If the T1-weighted image was acquired multi-slice (with slice
selection), rather than 3-D (with the slice dimension being
phase-encoded), then select this option.
 slider at its
default value of 5. This should not need to be changed.
slider at its
default value of 5. This should not need to be changed.
 button to start the
analysis, which will take some time. When the analysis is complete, you will
see a pop-up message showing the results:
button to start the
analysis, which will take some time. When the analysis is complete, you will
see a pop-up message showing the results:

