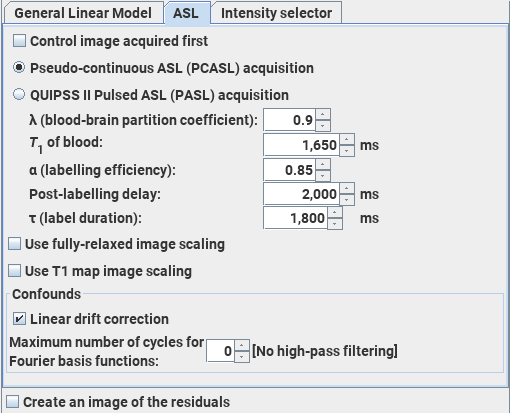
The Arterial Spin-Labelling module performs an analysis of ASL data acquired using alternating spin-labelled/control MR images to produce maps of cerebral blood flow (CBF). The analysis follows that recommended in: Magn Reson Med.73:102-16 (2015) "Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia." Alsop D.C. et al. The default values for the settings below come from this paper.
You can analyse images acquired using either pseudo-continuous ASL (PCASL) or QUIPSS II pulsed ASL (PASL) to obtain perfusion values. The units of CBF in the output image are mL/(100 g)/minute.
Note: the ability to obtain meaningful quantitative perfusion values is heavily-dependent on the implementation of the ASL pulse sequence and entering accurate values for the sequence parameters required below. Obtaining an accurate estimate of the labelling efficiency is particularly difficult, and poor estimates will lead to systematic errors in the perfusion values.
You choose the Arterial Spin-Labelling module by clicking the ASL tab in the Dynamic Analysis tool.

Set up the input images as detailed in the Introduction, noting that the number of time-points should be set equal to sum the number of labelled plus control images (i.e., twice the number of labelled images).
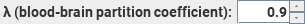 This
sets the partition coefficient for water in exchange between the blood and
the brain parenchyma. 0.9 is the recommended setting.
This
sets the partition coefficient for water in exchange between the blood and
the brain parenchyma. 0.9 is the recommended setting.
 This
sets the longitudinal relaxation time (T1) of whole blood.
The default of 1,650 ms is the value at 3.0 Tesla. At 1.5
Tesla, the T1 of blood is approximately 1350 ms. Set the
value according to the field strength of your MRI scanner.
This
sets the longitudinal relaxation time (T1) of whole blood.
The default of 1,650 ms is the value at 3.0 Tesla. At 1.5
Tesla, the T1 of blood is approximately 1350 ms. Set the
value according to the field strength of your MRI scanner.
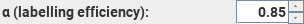 This
sets the labelling efficiency of the pulse sequence used.
The default value of 0.85 is that expected for PCASL; for PASL the expected
value is 0.98. However, these values are likely to be very implementation-dependent.
This
sets the labelling efficiency of the pulse sequence used.
The default value of 0.85 is that expected for PCASL; for PASL the expected
value is 0.98. However, these values are likely to be very implementation-dependent.
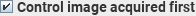 Normally, the first image acquired is a labelled
image, followed by a control image. Labelled and control images are
acquired repeatedly in the same order. If the first image acquired was the
control image, then select this checkbox.
Normally, the first image acquired is a labelled
image, followed by a control image. Labelled and control images are
acquired repeatedly in the same order. If the first image acquired was the
control image, then select this checkbox.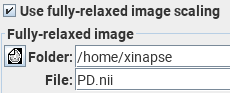
If you don't have such a PD image, then Jim will do a semi-quantitative analysis by using an average of all the "control" images as a substitute for the PD image. As long as the repetition time (TR) is very long compared to the T1 relaxation times, this this may be a reasonable approximation.
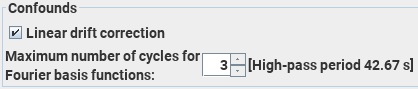
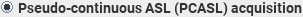 . Then set the additional parameters:
. Then set the additional parameters:


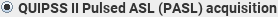 . Then set the additional parameters:
. Then set the additional parameters:


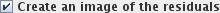 check-box will create
output image(s) with the same names as the input image(s), but with the suffix "residuals". The
residuals image(s) will be the result of subtracting the modelled ASL signal
(including the confounds) from the input data. Residuals images
can be useful for investigating whether fitting the ASL still leaves some "structure" - this would
be the case if the ASL does not provide a compete description of the input images, and that
possibly further correlates should be included in the model. In the case of a "complete" ASL, the
residuals will consist entirely of Gaussian-distributed uncorrelated values.
check-box will create
output image(s) with the same names as the input image(s), but with the suffix "residuals". The
residuals image(s) will be the result of subtracting the modelled ASL signal
(including the confounds) from the input data. Residuals images
can be useful for investigating whether fitting the ASL still leaves some "structure" - this would
be the case if the ASL does not provide a compete description of the input images, and that
possibly further correlates should be included in the model. In the case of a "complete" ASL, the
residuals will consist entirely of Gaussian-distributed uncorrelated values.
 button, the output image
created will have a name produced by taking the base name in the image output section and appending the
suffix "CBF". The units of CBF in the output image are mL/(100 g)/minute.
button, the output image
created will have a name produced by taking the base name in the image output section and appending the
suffix "CBF". The units of CBF in the output image are mL/(100 g)/minute.