- Place each subject's T1-weighted and a FLAIR image in its own folder.
- Rename the T1-weighted and a FLAIR image to have some consistency across all subjects. For example, you could name them "t1.nii" and "flair.nii".
 . Set a Basename to use for the
registered prior image, for example "Training".
Now press the
. Set a Basename to use for the
registered prior image, for example "Training".
Now press the
 button, which will perform
all image registrations needed.
button, which will perform
all image registrations needed.
Load the registered FLAIR image into Jim's main display, and launch the ROI Toolkit. Use Contoured, Irregular ROIs to outline the WM hyperintensities on every slice. Then save the ROIs to file, using the default file name (i.e., if your registered FLAIR image is called "rflair.nii", then the ROI file name must be "rflair.roi").
Repeat for all subjects in your training set. The number of subjects needed for training will vary according to their lesion loads, but we suggest that at least 20 subjects will be needed, and possibly more.
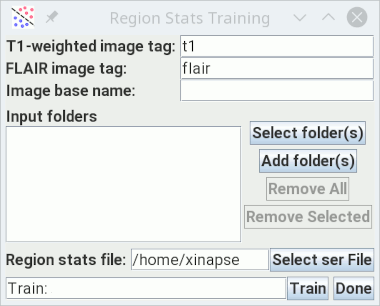
- Name tags for the T1-weighted and FLAIR images. The image names for every subject must contain these tags. For example, you could have a series of patients, each in their own separate folder, with name "Pat1t1.nii" & "Pat1flair.nii", "Pat2t1.nii" & "Pat2flair.nii" etc.
- The image Basename. This must be the same Basename that you used in step 1 (e.g., "Training").
- Select all the folders that contain the training subjects' images and ROI file. There will be one folder for each training subject. You can select a single "top-level" folder if all the different subjects are within this folder.
- Set the name of the file to which you want to save the region statistics. This should have a file extension ".ser".
 button. Training will
start, processing each training subject in turn. When, complete, the
region statistics file will be created, and can be used as input
to the atrophy assessment with WM lesions, either
cross-sectional or
longitudinal.
button. Training will
start, processing each training subject in turn. When, complete, the
region statistics file will be created, and can be used as input
to the atrophy assessment with WM lesions, either
cross-sectional or
longitudinal.